Core-Developer
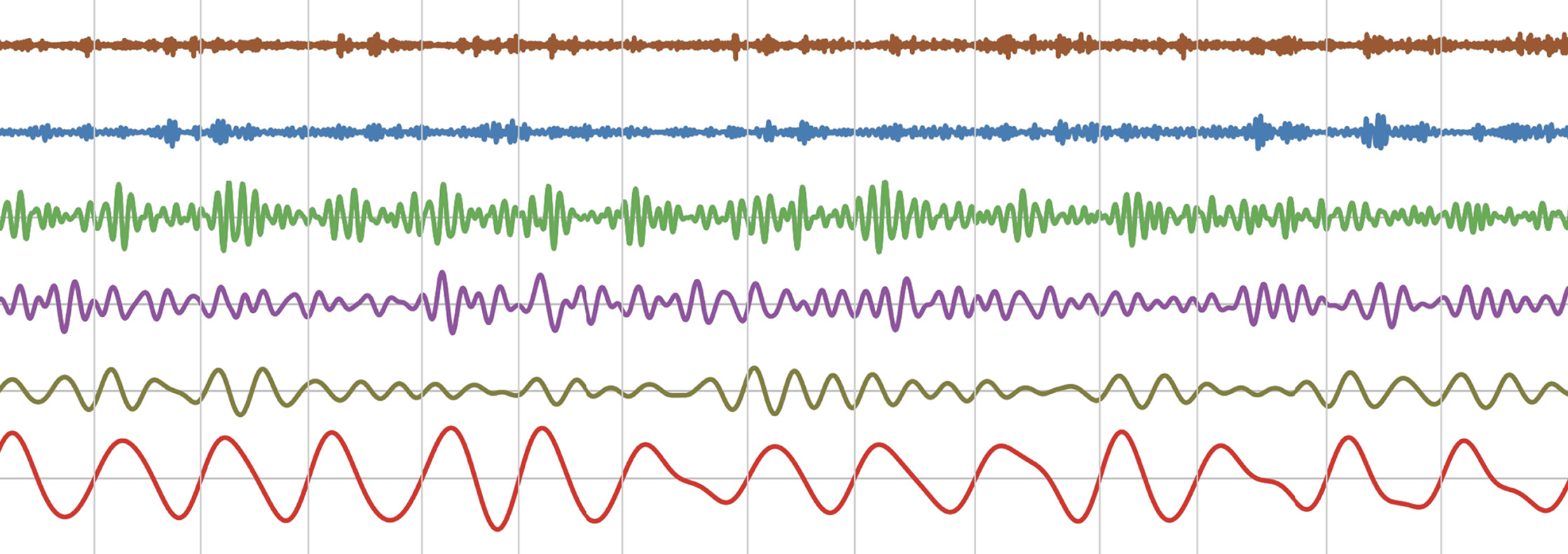
EMD: Empirical Mode Decomposition
The Empirical Mode Decomposition (EMD) package contains Python functions for analysis of non-linear and non-stationary oscillatory time series. EMD implements a family of sifting algorithms, instantaneous frequency transformations, power spectrum construction and single-cycle feature extraction.





Andrew Quinn, Vitor Lopes-dos-Santos, David Dupret, Anna Nobre & Mark Woolrich (Mar 2021)
The Journal of Open Source Software doi
Andrew J. Quinn, Vítor Lopes-dos-Santos, Norden Huang, Wei-Kuang Liang, Chi-Hung Juan, Jia-Rong Yeh, Anna C. Nobre, David Dupret & Mark W. Woolrich (Apr 2021)
bioRxiv (Cold Spring Harbor Laboratory) doi
Marco S. Fabus, Mark W. Woolrich, Catherine W. Warnaby & Andrew J. Quinn (Jan 2022)
IEEE Open Journal of Signal Processing doi
Marco S. Fabus, Andrew J. Quinn, Catherine E. Warnaby & Mark W. Woolrich (Jul 2021)
bioRxiv (Cold Spring Harbor Laboratory) doi
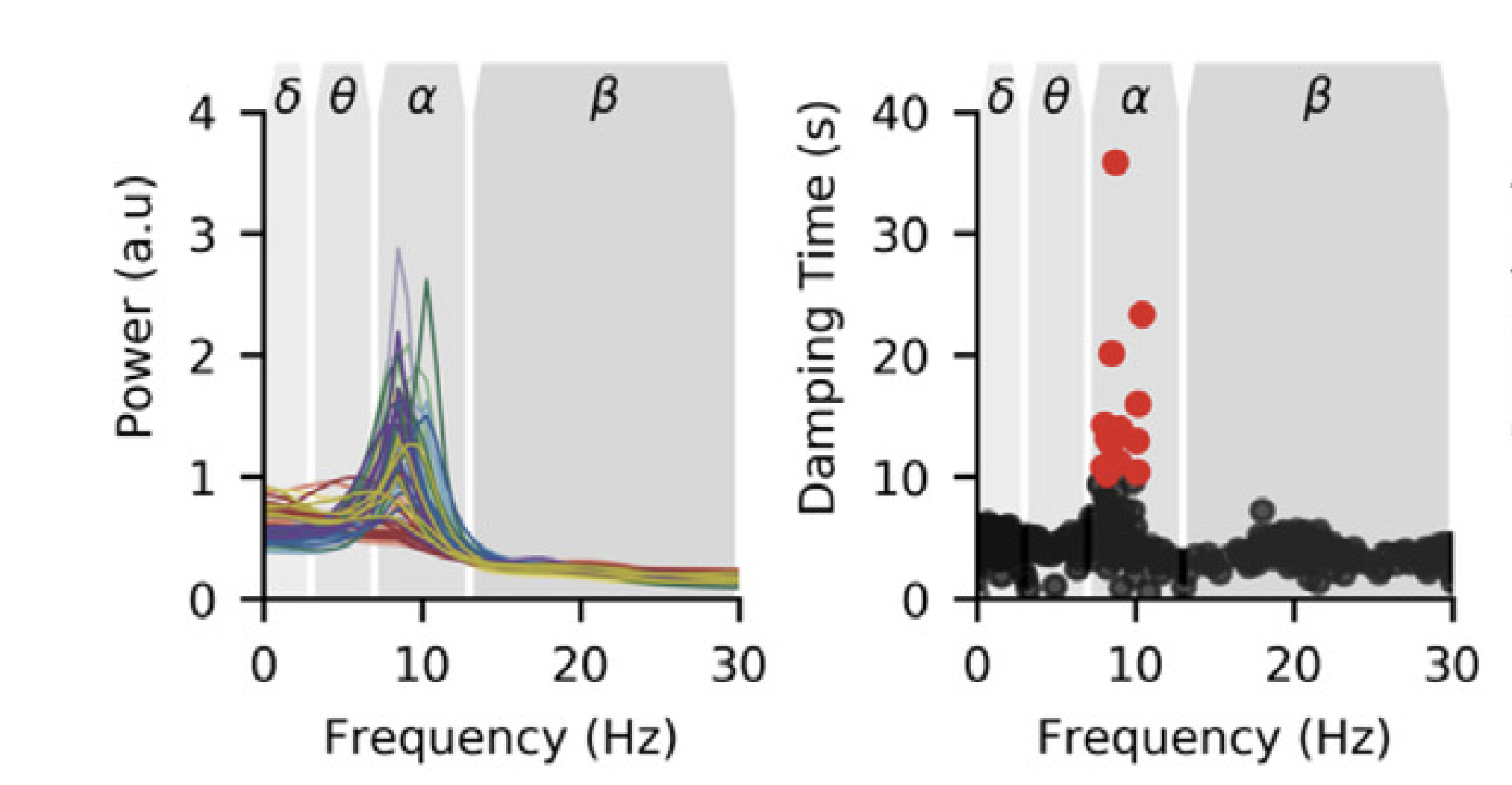
SAILS: Spectrum Analysis in Linear Systems
SAILS is a python package autoregressive modelling of mutlltivariate time series. Routines are provided for linear model fitting, model validation, power spectrum estimation and connectivity estimation. SAILS also provides functionality for decomposing fitted autoregressive models into a set of data-driven multivariate oscillatory modes.





Andrew Quinn & Mark Hymers (Mar 2020)
The Journal of Open Source Software doi
Andrew J. Quinn, Gary G.R. Green & Mark Hymers (Jul 2021)
NeuroImage doi
Andrew J Quinn, Lauren Z Atkinson, Chetan Gohil, Oliver Kohl, Jemma Pitt, Catharina Zich, Anna C Nobre & Mark W Woolrich (Nov 2022)
bioRxiv (Cold Spring Harbor Laboratory) doi
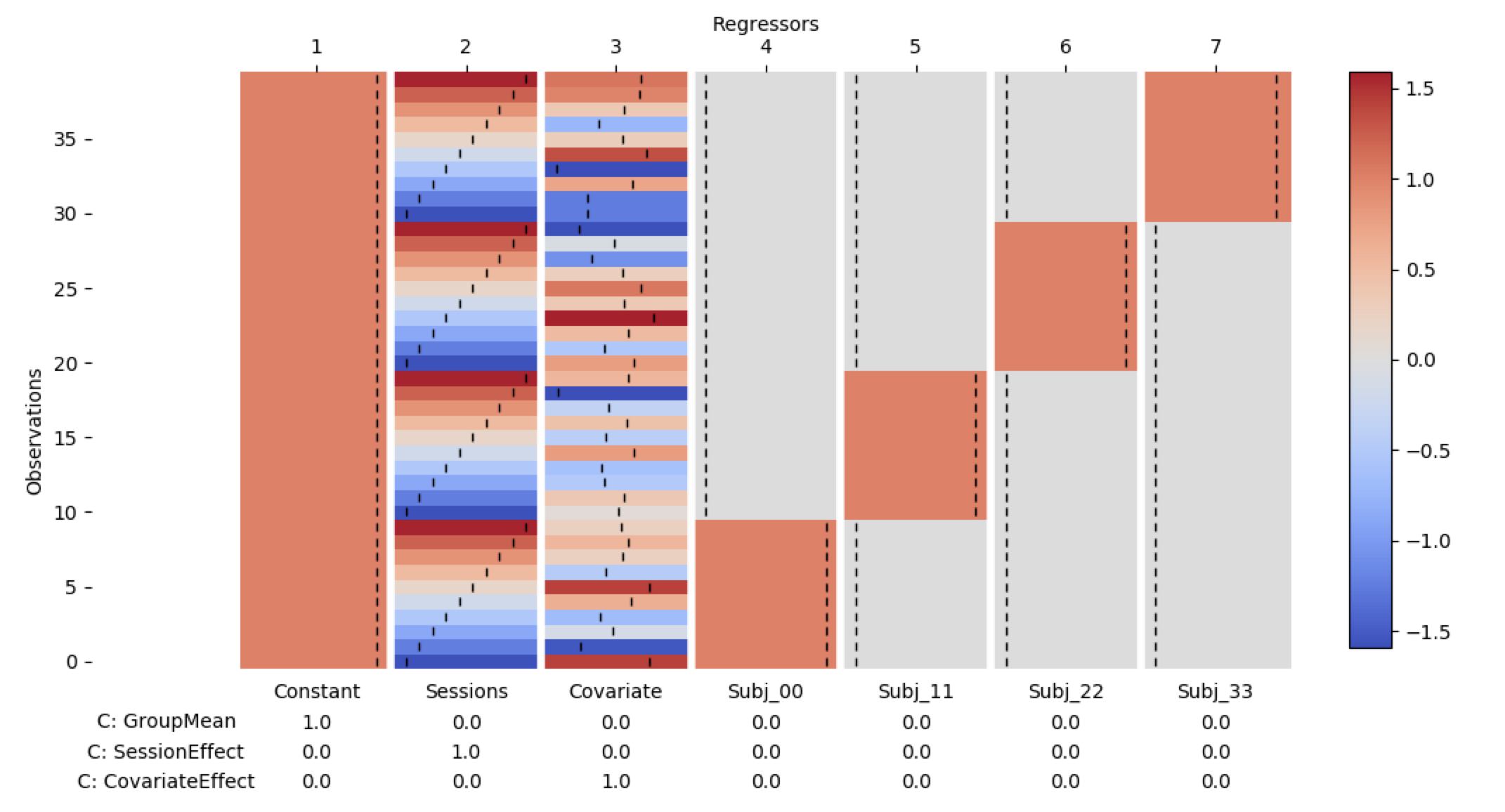
GLMTools
glmtools is a Python package which aims to simplify the definition, creation and reuse of design matrices for General Linear Models. It contains routines for abstractly defining a GLM design before applying this definition to new data to create a design matrix. There are routines for basic linear model fitting, t-stat calculation and non-parametric permutations.





Andrew J Quinn, Lauren Z Atkinson, Chetan Gohil, Oliver Kohl, Jemma Pitt, Catharina Zich, Anna C Nobre & Mark W Woolrich (Nov 2022)
bioRxiv (Cold Spring Harbor Laboratory) doi
Alexander L Anwyl-Irvine, Edwin S Dalmaijer, Andrew J Quinn, Amy Johnson & Duncan E Astle (Dec 2020)
Cerebral Cortex Communications doi
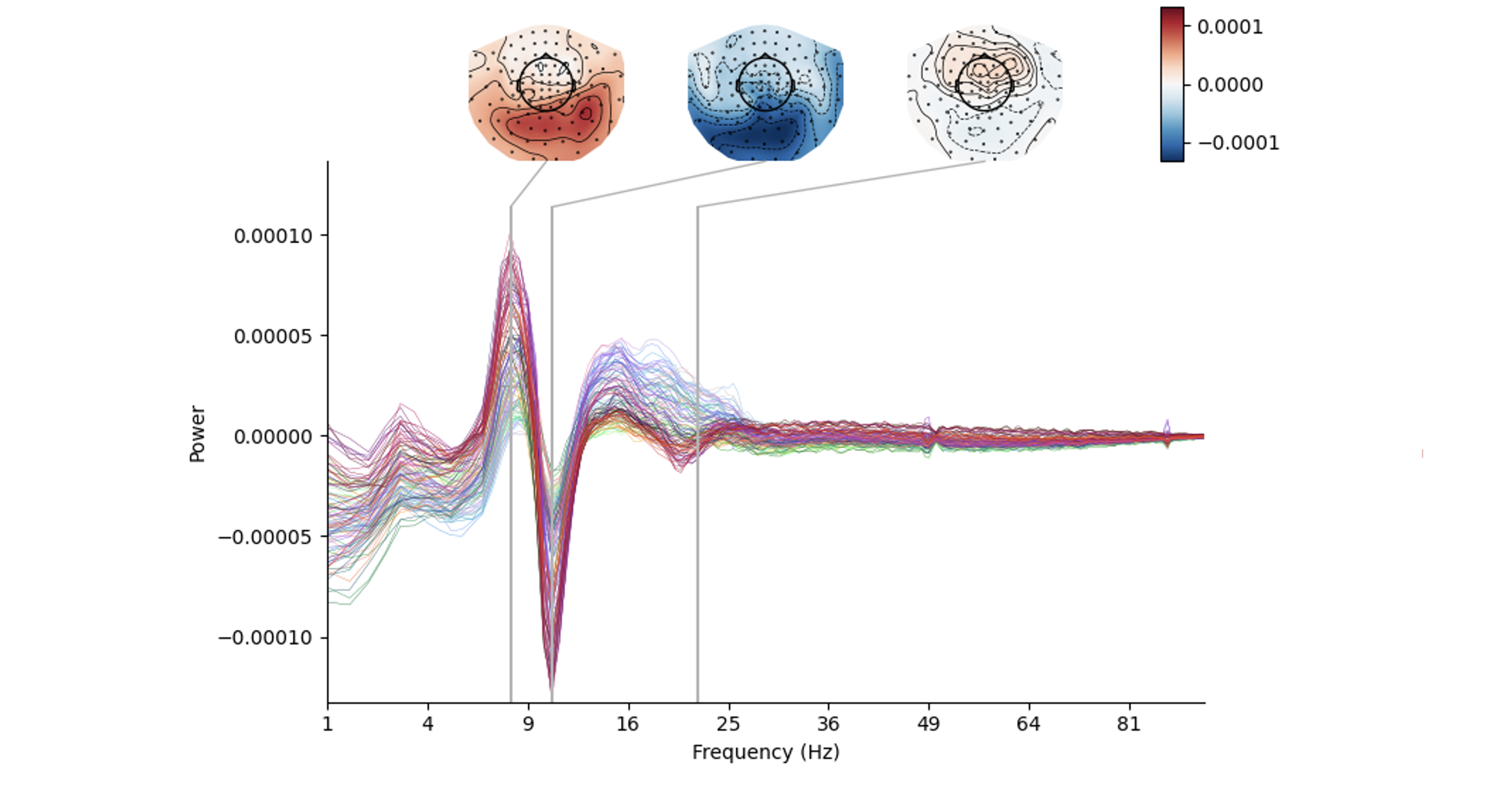
Andrew J Quinn, Lauren Z Atkinson, Chetan Gohil, Oliver Kohl, Jemma Pitt, Catharina Zich, Anna C Nobre & Mark W Woolrich (Nov 2022)
bioRxiv (Cold Spring Harbor Laboratory) doi



